PAN-GENOME: UNLOCKING THE POTENTIAL OF DIVERSITY
Over the next 30 years, the Earth’s population is projected to grow from the present 7.8 billion to 9.7 billion. In parallel, climate change will bring with it major changes and shifts in rainfall, temperature and rising sea levels, placing already at risk regions into jeopardy. We are already beginning to face the phenomenon of climate refugees around the world, as populations can no longer maintain a living where they currently live and are forced to migrate elsewhere.
What are some of our options? Mitigation of these challenges will require rapid adaption of germplasm in current and emerging crops. Advances in crop breeding have been accepted as a “genetic gain” with a fraction of enhancement in yield potential. Consecutive to intensive selection and inbreeding depression, the genetic pool suffered significantly due to narrow genetic diversity. Necessity is the mother of invention thus the rapid enhancement of genomics and high throughput experimentation are now broadening the funnel and enhancing the scope of breeding. This has led to a better selection of germplasm from wild relatives and prediction of adaptive complex traits in new environments.
What about the future? Next-generation agricultural breeding strategies must produce more yield consistently under changing environments and bring more value to farm income.
“The value and utility of any experiment are determined by the fitness of the material to the purpose for which it is used, and thus in the case before us it cannot be immaterial what plants are subjected to experiment and in what manner such experiment is conducted.”
Genetic diversity provides the base for selection in a given population, created on allele frequencies. Too much diversity was not considered as an option to broaden the genetic pool to avoid the cost associated of bringing those alleles in core gene pool. In addition, there was a lack of understating of the genomic content due to limitations of the reference genome.
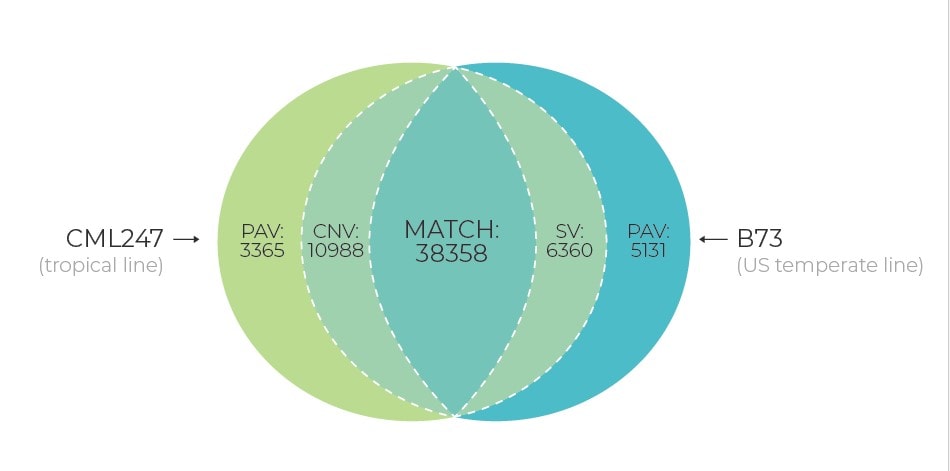
Presently, with the increasing number of whole genome projects, there are evidences that a single reference sequence is inadequate to capture genetic diversity within a whole species due to the presence of significant levels of structural variation, copy number variants (CNVs), presence/absence variants (PAVs), and more events of translocation and inversions. To understand these genetic variations within a species and the collective impact on different traits/attributes, it’s important to construct a pan-genome approach. Pan-genome will represent the complete set of genes for a given species by including genetically diverse widespread lines. Characterization of these pan-genomes will lead to the transformation of our knowledge and provide a holistic approach to understand the genetic diversity of crops.
In the recent publication by Bayer et al. (2020), they stated that:
“Pan-Genome studies will bring an understanding on the impact of variable genes as a “new reference” for a given crop species. This will also provide a broad insight into evolution, selection and in particular the functionality of genomes.”
A pan-genome structure has multiple advantages over a single, linear reference genome sequence in plant breeding applications. With a pan-genome available for a given crop that includes diverse germplasm, it will provide a single coordinate system to anchor all known variations and phenotype information, and will allow for the identification of novel genes from the available germplasm that are not present in the single reference genome. Moreover, the pan-genome will reveal chromosomal rearrangements between genotypes that can hinder the introgression of desired genes via re-synthesis or crossing of elite material to more diverse landrace or wild germplasm.
The use of a pan-genome representing the complete gene content of any given species, together with the latest in high-throughput genome sequencing would increase the efficiency of SNP calling in several ways and allow the technology to play a growing central role in breeding.
The generation of a canola, tomato, cotton and pepper pan-genome is already underway. In the case of canola, effective genomics strategies have been proposed by the Canola Council of Canada to help canola producers improve crop yield and support the industry’s efforts to increase average yield to 52 bus/ac by 2025. Currently, there is only one public reference available for canola, which is the European winter variety.

NRGene now has all-to-all linear mapping of multiple de-novo assemblies to enable intra-species gene content variation exploration and has established pan-genome consortium projects to capture the broad genetic diversity of a specific crop. Each project is organized under public-private partnership.
Irrespective of the algorithm used, or the approach employed to identify SNPs and structural variation, using a pan-genome as the reference for read mapping would consider regions displaying complex variation. In this way, it will be possible to know the number and types of variants contributed by each individual in the pan-genome. This would also increase the overall number of variants that would be identified, if the reference had otherwise been based on a single individual alone. In addition, using the pan-genome as a basis for SNP discovery would cut down the time/cost and effort required to map reads to several individuals one at a time.
The future is looking better for both the agricultural and scientific communities.
Ask the author
We want to hear more about your needs. Please fill the form below and member of our team will contact you in the next few days.